NEAT was invented to solve the competing conventions problem.
… by figuring out how to take two different parental neural networks, and identify parts that are likely to be performing similar functions. It does so by tracking the ancestry of edges (nature does things akin to this too).
Grows networks from simple to complex (Complexification).
Avoids searching unnecessariy high-d spaces.
Terminology
Genotype = “blueprint”
Phenotype = actual structure
NeuroEvolution of Augmenting Topologies = evolving the structure (topology) of the network
Intro example
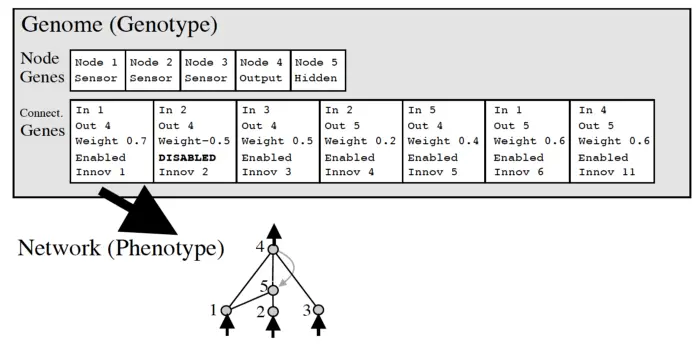
Here, the genotype is encoded as two vectors, one for the nodes and the connectivity.
The network currently has 5 neurons, each with a flag indicating neuron type.
There are six synapses, however one of them is disabled, so the phenotype only has 5 synapses.
The disabled flag mimicks dominant / recessive genes. Intuitively, the connection might become useful in a different context at a later point in time. In nature, this is sometimes called “junk DNA”, genes that are there but not active. It is like a scratchpad / like uncommenting out code.
We also have an innovation number, the main innovation of NEAT.
Innovation number
The innovation number is a globally unique counter “historical marking”, assigned to each new node / synapse separately. They identify the genes.
Nodes with the same innovation number likely perform similar functions, kind of like alleles (different version of the same gene).
Mutation
One parent → One child.
Copy parent weights (retain innovation number).
Add / disable (never remove) nodes/synapses and change weights (always).
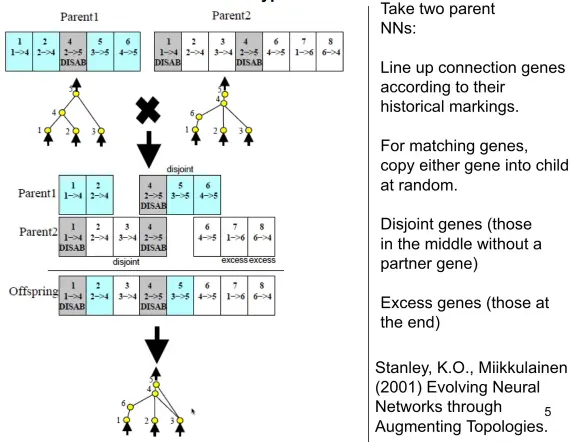
Crossover
Produces children that are similar to their parents.
Procedure:
Align genes by innovaiton number.
For each shared gene, flip a coin to decide which parent to inherit from.
For disjoint and excess genes are inherited from the fitter parent. If they are equally fit, flip a coin to decide whether to inherit it.
A disjoint gene is one that is present in one parent but not the other. Excess genes are disjoint genes… just where we ran out of innovation numbers from the other parent.
Champion
The best out of each species is kept in the population, unmodified, if the species has more than 5 members.
Why preserve champions? Fitness can only go up or flat, prevents catastrophic loss due to bad mutation/crossover, exploration vs exploitation: The champion exploits known good solutions while its offspring explore variations.
Link to originalComment by Kenneth O. Stanley on discussion about historical markers for nodes vs connections (a divergence of the tensorNEAT implementation from original NEAT):
TLDR h/t claude:
The fundamental difference centers on historical markers (innovation numbers):
- Original NEAT: Places unique historical markers on every connection (edge) in the network
- TensorNEAT/Python-NEAT: Only places historical markers on nodes, using (start_node, end_node) tuples to identify connections
Implications:
- Homology determination: When two genomes independently evolve the same connection (e.g., node 2→3), original NEAT treats them as different genes (disjoint), while TensorNEAT treats them as the same gene (matching)
- Crossover behavior: This affects which genes get inherited during reproduction - matching genes are selected 50/50 from parents, while disjoint genes come from the fitter parent
- Speciation: Distance calculations for species formation are computed differently, potentially creating more inclusive species in TensorNEAT
Performance Impact
- The difference likely has some adverse effect on efficiency, but probably not very large
- The main risk is wasting effort crossing over incompatible individuals from distant lineages
- However, speciation should naturally keep most truly distant genomes apart anyway
- The impact would mainly affect “borderline cases” at the edge of compatibility
- Empirical testing would be needed to determine the actual performance difference
Philosophical Perspective
- From a biological standpoint, genes from very distant lineages shouldn’t be considered “the same” just because they’re structurally similar
- His analogy: “if a fish had a mutation that encodes the same protein as something in the human gut, I would not regard the fish as being more compatible with a human for the purposes of mating”
- He estimates “while there may be some difference in performance, it would likely not be very large”
Resolution:
- WLS2002 implemented
OriginNodeandOriginConnclasses in TensorNEAT that follow the original NEAT paper’s approach- Initial experiments by sopotc showed 10-15% better fitness with the original implementation, though more rigorous testing is needed
Initialization
NEAT biases the search towards minimal-dimensional spaces by starting out with a uniform population of networks with zero hidden nodes (i.e., all inputs connect directly to outputs).
New structure is introduced incrementally as structural mutations occur, and only those structures survive that are found to be useful through fitness evaluations. In other words, the structural elaborations that occur in NEAT are always justified.
The population is divided into clusters based on their similarity, and measures are put in place from a single species dominating, to keep diversity over time (protecting innovation).
TODO details (for now, see my implementation: https://github.com/MaxWolf-01/neat
Fitness sharing function: Uses a binary “1 if same species 0 if other” which reduces to dividing individual’s fitness by number of members in the species.
Ig non-binary fitness would be implemented with similarity between species, s.t. similar species also share fitness, thereby increasing pressure on diversity even more.
From the NEAT paper:
… because indirect encoding do not map directly to their phenotypes, they can bias the search in unpredictable ways. To make good use of indirect encodings, we need to first understand them well enough to make sure that they do not focus the search on some suboptimal class of topologies
References
(ES-(Hyper-))NEAT resources
Pytoch (Hyper)NEAT code
HyperNEAT paper (high quality)
Good BP series:Could not find ES-HyperNEAT paper… There is “Enhancing ES-HyperNEAT” (same authors).
→ nvm, found it. See ES-HyperNEAT.
Bad ES-HyperNEAT code
Evolutionary Robotics, Lecture 16: NEAT & HyperNEAT (Josh Bongard)
ES-HyperNEAT
evolutionary optimization
neuroevolution
Kenneth O. Stanley
neural architecture search