year: 2023/07
paper: https://arxiv.org/pdf/2307.08197.pdf
website: NDP summary - YT !
code: https://github.com/enajx/NDP/tree/main | https://github.com/enajx/NDP/tree/main/NDP-RL
connections: neuroevolution, ITU Copenhagen, indirect encoding, developmental encoding
youtube summary AT ALIFE2023: Towards Self-Assembling Artificial Neural Networks through Neural Developmental Programs
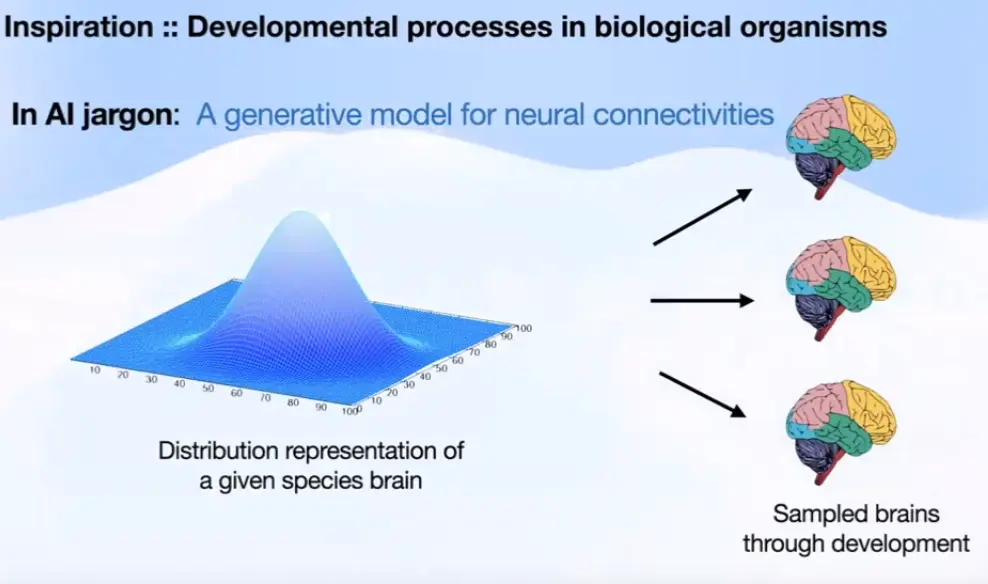
Link to originalHow do biological organisms grow their nerve systems?
Neural circutry is grown through a developmental process.
Each cell contains the same developmental program.Indirect encodings:
Developmental indirect encodings:
→ Can’t arrive at the phenotype in a single time-step, but unfold it through a series of time-steps.
Conjectured to be analogous to computational irreducibility in some CA rules.What’s interesting about indirect encodings?
compression: size of genotype << size of phenotype (1GB << 700TB)
generalization: genomic bottleneck hypothesis (incentivized to learn adaptive behaviors)
It’s easier to find a program that generates a complex structure than to directly search the space of complex structures.
Start with minimal initial network → Graph convolution aggregates information over states → growth network decides whether to add node / connection → …
Difference to NEAT: NDPs grow structure not during evolutionary time, but during developmental time.
NDP is a generalization of NCA to graph structures.
LNDP introduces life-time plasticity. → LNDP also gets the reward as input!
To avoid collapse of these systems: lateral inhibition + diversity
They’re hard to train → combining it with open-ended curricula might let this approach unfold its full potential